NetDecoder is developed by Hu Li's Lab. It is a novel network biology-based computational platform designed to integrate transcriptomes, interactomes and gene ontologies to identify phenotype-specific subnetworks. NetDecoder is based on network flow algorithm and formulated as a minimum-cost flow optimization problem to identify and prioritize paths and key regulators within disease specific subnetworks. NetDecoder is designed to capture molecular switches and infer disease-specific networks to better understand pathways and identify key regulators that contribute to a disease phenotype. NetDecoder has extensive documentation and tutorial with free software package downloadable for the research communities. You can use NetDecoder on-line by uploading your data here, or you can download and run NetDecoder locally on your computer. NetDecoder is provided under the OSI-approved Artistic License (version 2.0).
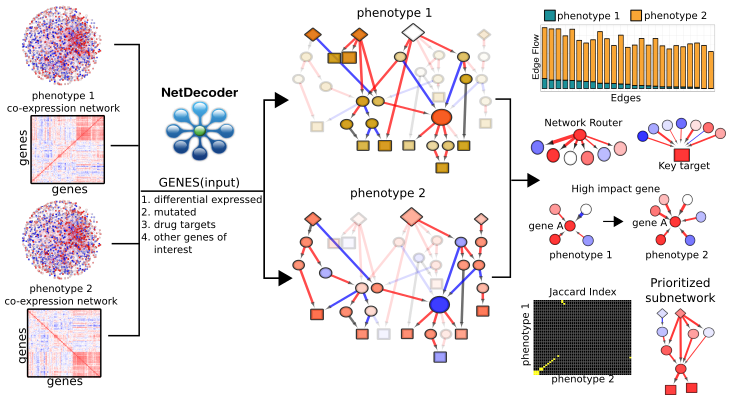
NetDecoder takes as input a phenotype-specific edge-weighted interaction network and a list of genes to be used as sources and returns:
- Prioritized gene-gene interactions. These interactions are edges with higher flows under phenotype 2 than in phenotype 1. Typically, phenotype 1 represents the transcriptome of the “control” samples while phenotype 2 represents the transcriptome of a disease state.
- Intermediary genes with high flow differences between phenotype 1 and phenotype 2 subnetworks. These genes are called network routers.
- Key targets, which are transcription-related genes with high flow differences between phenotype 1 and phenotype 2 subnetworks.
- Impact scores for all intermediary and sink (target) genes reflecting the extent to which these genes have changed between phenotype 1 and phenotype 2 subnetworks.
- Evaluation of network similarity between phenotype 1 and phenotype 2 subnetworks.
- Prioritized and edge-centered subnetworks.